Jun 10 2016
Knowing the exact number of molecules located at specific junctures in cells can be a critical measure of health as well as disease. For example, abnormally high numbers of growth factor receptors on cells can be an indication of cancerous and precancerous states; specific proteins located at the junction where neurons connect in the brain may affect brain function as they accumulate or disperse.
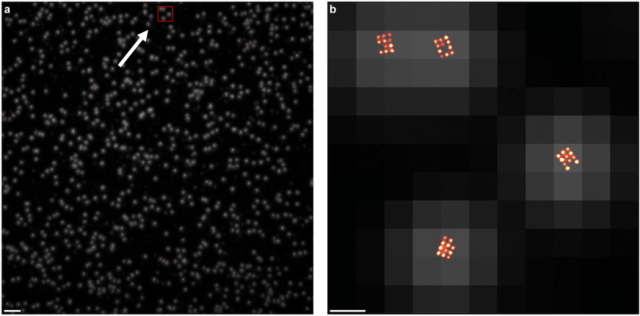 What appear to be single dots of light in the left panel (taken at low resolution) are actually grids of 11 individually labeled molecules, which can be seen at high resolution, in the right panel. Right panel shows the area of detail noted by the arrow and red box in the left. The ability to pinpoint exact numbers and distributions of molecules in cells can reveal vital health information. (Source: Macmillan Publishers)
What appear to be single dots of light in the left panel (taken at low resolution) are actually grids of 11 individually labeled molecules, which can be seen at high resolution, in the right panel. Right panel shows the area of detail noted by the arrow and red box in the left. The ability to pinpoint exact numbers and distributions of molecules in cells can reveal vital health information. (Source: Macmillan Publishers)
Until recently, researchers have had to use either very expensive microscope hardware, or highly complex -- and often imprecise -- microscopy software, to see individual, fluorescently-labeled molecules in tightly bunched groups in cells. Now, a simplified method known as qPAINT uses the blinking pattern of the light that marks each molecule, to find, count and study individual molecules that are just a few nanometers apart -- all using the standard microscopes already found in laboratories.
“qPAINT allows identification of each point of light coming from a labeled molecule without the need for complex and sometimes inexact microscopy calculations,” explains Behrouz Shabestari, Ph.D., Director of the NIBIB program in Optical Imaging and Spectroscopy. “The method overcomes the problem that occurs when trying to visualize molecular structures that are in very close proximity: light diffuses as it leaves the spot where it originates. This masks exactly how many points of light—each representing a single molecule—are actually creating the light.”
For example, if someone shines a flashlight at you, what you see is a beam of light much wider than the actual flashlight because the light spreads out, or diffuses, as it moves toward you. If the person was actually holding two or three flashlights in a bundle you would still see one diffuse beam of light and would not be able to tell how many flashlights they were holding.
Researchers at the Wyss Institute for Biologically Inspired Engineering at Harvard University have developed a technique that solves this problem and allows the identification of individual, fluorescently- labeled molecules even if they are right next to each other. The method is reported in the March 28 issue of Nature Methods.1
The new qPAINT technology builds on the group’s DNA-PAINT technology, which uses fluorescent probes to find their targets using DNA binding. The qPAINT system takes advantage of the fact that the fluorescent DNA probes bind to and then detach from the target at a specific rate. When bound to its target, the fluorescent dye lights up; when detached, the light turns off. This causes a blinking pattern the qPAINT technology detects and translates to reveal exactly how many molecules are in a group in a specific location of interest in the cell.
For example, if there is just one target molecule in the location of interest in the cell, the fluorescent probe might bind and release from that target two times every second. So, qPAINT detects two blinks per second, which tells the researcher there is one target molecule at that location. If there are two molecules at that location, qPAINT would detect four blinks per second (two for each molecule.) If there were three molecules qPAINT would detect six blinks per second--two blinks being made by each of the three target molecules.
“The critical point here,” explains Peng Yin, Ph.D., Core Faculty Member at the Wyss Institute and senior author of the study, “is that very expensive, super-resolution microscopes can actually resolve the diffuse light coming from multiple targets in close proximity and allow one to see the individual targets. But, few laboratories can afford these expensive microscopes. Our qPAINT technology reveals the same precise information using microscopes that are ten times less expensive and are already standard equipment in most laboratories. qPAINT can also detect these targets at deeper depths in tissues than is capable with the very expensive microscopes. It is also much simpler and more precise than other software-based methods that are currently used with less expensive microscopes.”
The team’s goal is to develop technologies that will allow the majority of research labs to access the most powerful techniques. Their most recent effort is a start-up company that will make qPAINT commercially available. They are also developing an even more economical microscope than is available currently, to work with qPAINT.
The start-up company is called Ultivue, and aims to provide “the ultimate view into biology,” says Yin. “We see qPAINT and Ultivue as agents for research democratization,” Yin adds, referring to making powerful technologies affordable and accessible to as many researchers as possible.
The work was supported by grant EB018659 from the National Institute of Biomedical Imaging and Bioengineering and a National Institutes of Health New Innovator Award OD007292. Additional funding was provided by the National Science Foundation and the Office of Naval Research.