Mar 24 2017
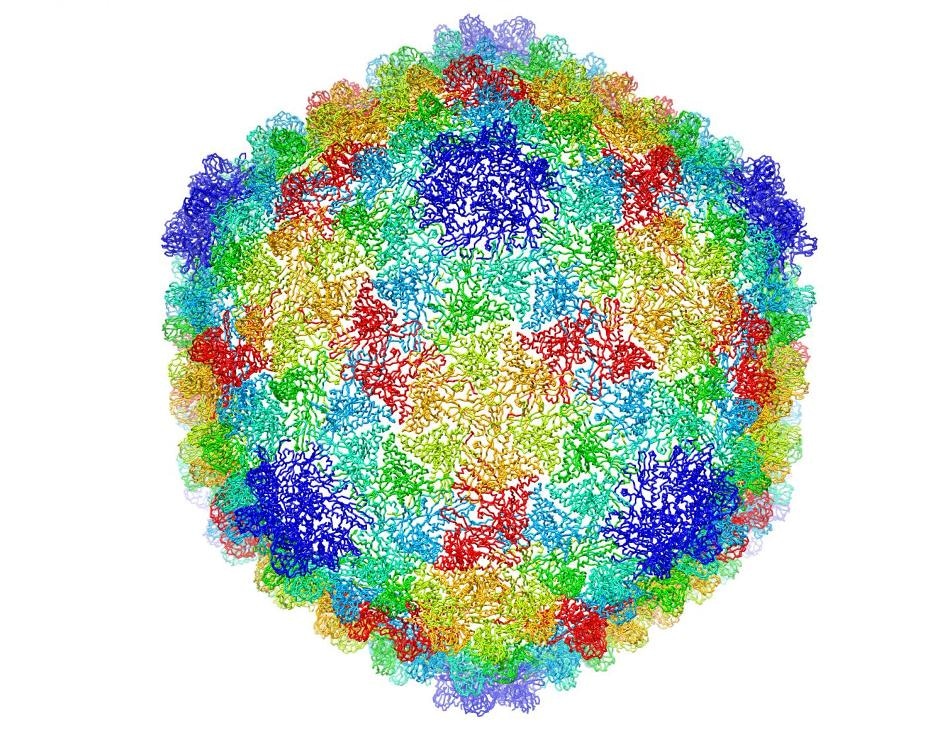 Complete capsid of bacteriophage P22 generated with validated atomic models that were derived from a high-resolution cryo-electron microscopy density map. (C. Hryc and the Chiu Lab, Baylor College of Medicine)
Complete capsid of bacteriophage P22 generated with validated atomic models that were derived from a high-resolution cryo-electron microscopy density map. (C. Hryc and the Chiu Lab, Baylor College of Medicine)
Cryo-electron microscopy (cryo-EM), which enables the molecular level visualization of proteins, viruses, and other biological structures, is an important tool used to further biochemical knowledge.
Recently, researchers from Lawrence Berkeley National Laboratory (Berkeley Lab) have broadened the cryo-EM’s impact by formulating a new computational algorithm that was helpful in building a 3D atomic-scale model of bacteriophage P22 for the first time.
More than 20,000 2D cryo-EM images of bacteriophage P22 (also referred to as the P22 virus that infects the common bacterium Salmonella) from Baylor College of Medicine were used to create the model. The researchers from Baylor College of Medicine, Massachusetts Institute of Technology, Purdue University and Berkeley Lab published the results in the Proceedings of the National Academies of Sciences earlier in March.
This is a great example of how to exploit electron microscopy technology and combine it with new computational methods to determine a bacteriophage’s structure. We developed the algorithms—the computational code—to optimize the atomic model so that it best fit the experimental data.
Paul Adams, Berkeley Lab’s Molecular Biophysics & Integrated Bioimaging Division Director
Pavel Afonine, a Berkeley Lab computational research scientist and the paper’s co-author, took the initiative in formulating the algorithm using Phenix, a software suite used customarily in X-ray crystallography for establishing macromolecular structures.
The successful version of bacteriophage P22’s 3D atomic-scale model permits scientists to see inside the virus’ protein coats at resolution. It is the result of numerous years of research that formerly had enabled Baylor College researchers to trace out a majority of the protein’s backbone, but not the fine details, according to Corey Hryc, co-first author and a graduate student of Baylor biochemistry professor Wah Chiu.
Thanks to this exquisite structural detail, we have determined the protein chemistry of the P22 virus. I think it is important that we provide detailed annotations with the structure so other researchers can use it for their future experiments.
Wah Chiu, Professor, Berkeley Lab
Chiu’s lab has been using cryo-EM and computer reconstruction methods to construct 3D molecular structures for nearly three decades.
The findings are likely to have valuable biological repercussions as well.
Due to the 3D atomic-scale model, it is currently “possible to see the interactions between the pieces making up the P22 virus, which are critical to making it stable,” Adams said. This helps researchers to discover ways to make chemicals that can stick to specific proteins. Adams emphasizes that the ability to understand the atom configuration in molecular space can be used to produce new insights into drug design and development.
This research was funded by the National Institutes of Health.