Intrinsically disordered proteins (IDPs) play significant roles in various biological events. Due to their non-intuitive physical structures, analysis and understanding of their dynamics have proven challenging. Innovative atomic force microscopy techniques have emerged as a potential tool to gain insights into the structure of IDPs.

Image Credit: Elizaveta Galitckaia/Shutterstock.com
IDPs do not establish distinctive three-dimensional (3D) structures under natural circumstances. These novel proteins attract attention because, while not conforming to identifiable 3D structures, they can still perform various biological tasks.
Methods to Image Intrinsically Disordered Proteins
Since IDPs structure dynamically samples a wide range of conformational states, their structural study has been challenging.
IDPs cannot be effectively studied by X-ray crystallography or electron microscopy (EM) because they cannot be crystallized and are too thin to be seen by EM.
NMR is a relatively successful technique for the structural investigation of IDPs. Small-angle X-ray scattering (SAXS) is sometimes used to supplement NMR short-range structural data and estimate the overall dimensions of IDPs. Both are ensemble-averaging techniques, making it impossible to look at specific conformations and their populations.
For the structural characterization of chemically denatured proteins and IDPs, single-molecule Förster resonance energy transfer spectroscopy can be employed to resolve the dynamic diversity of specimen structure. However, the donor and acceptor must be close (less than 10 nm). To simultaneously and temporally examine the local and overall structures of IDPs, a gap in viable technology remains.
What is Atomic Force Microscopy (AFM)?
Throughout the history of imaging devices, microscopes have developed quickly in tandem with technological innovation, enabling high levels of detail in scientific research and material fabrication.
In 1981, Binning and Rohrer developed scanning tunneling microscopy (STM), a practical technique for examining the surface characteristics of materials. However, the requirements for electrical conductivity in STM samples limit the technology's potential uses.
Binning et al. developed the first AFM apparatus to circumvent this limitation. This advanced surface imaging technique, developed based on STM, uses the interaction between the probe and sample to examine the characteristics of material surfaces at the nanoscale.
A standard AFM includes a probe, laser, cantilever, electrically driven scanner, and photo-detector.
During scanning, the probe's movement in the x, y, and z axes is controlled by the piezoelectric module (PZT).
As the tip is scanned in the x and y directions, the surface undulation changes the tip's position in relation to the sample.
After the laser beam is directed at the back of the cantilever and reflected back to the photodetector, the difference in laser intensity is measured by the detector, which is proportional to the cantilever's deformation.
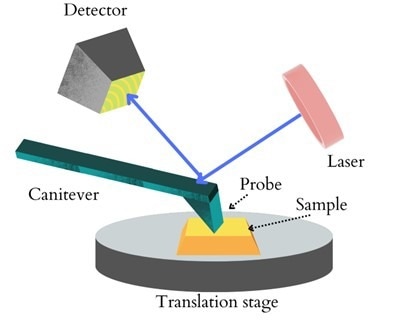
Figure 1: A schematic diagram of AFM. Image Credit: Ilamaran Sivarajah
The feedback system continually alters the distance between the tip and the sample based on fluctuations between detector signals to generate surface topography and phase maps.
Depending on the demands of the experiment, variations of the AFM have been regularly developed.
Specifically in structural biology, to overcome the limits imposed by light-based single-molecule biophysics, high-speed atomic force microscopy (HS-AFM) was created.
This technology makes it possible to directly observe single protein molecules in action at high spatiotemporal resolution.
High-Speed Atomic Force Microscopy
In a state-of-the-art HS-AFM, dimensions of the AFM cantilever are reduced, and the scanning speed is increased.
The cantilever’s resonant frequency is around 0.8-1.2 MHz with a spring constant of 0.1-0.2 N/m.
The fast scanner is operated with a resonant frequency of 100-170 kHz in the Z-direction. With this set-up, an HS-AFM can image protein molecules. The spatial resolution of HS-AFM is typically 2-3 nm in the lateral direction and 0.15 nm in the vertical direction.
Even at high bandwidth, the vertical resolution of HS-AFM is higher than that of slow AFM because the oscillation amplitude of shorter cantilevers can be detected more precisely and sensitively than that of typical long cantilevers. Unstructured polypeptides can be imaged with a vertical resolution of 0.15 nm.
The amplitude modulation mode is typically employed when using HS-AFM for IDP structural and dynamic research.
Amplitude modulation involves vertically oscillating a cantilever in buffer solution at its resonance frequency of about 1 MHz. The cantilever free oscillation amplitude and the set point amplitude are set at optimum values to regulate the loss of oscillation energy per tap.
IDRs' thin and flexible structure can be seen using HS-AFM.
Experiments have ruled out any potential impacts of the tip-sample contact on the structural characteristics and function of the protein under study. A Series of experiments on several IDPs demonstrate that HS-AFM can distinguish fully disordered regions in IDPs.
The Future of High-Speed Atomic Force Microscopy for Intrinsically Disordered Proteins
High-speed atomic force microscopy (HS-AFM) has been investigated as a potential method to describe the structure and behavior of IDPs.
Multiple HS-AFM scans of an IDP molecule can show disorder-to-order transitions and continuously folded and disordered regions of the molecule.
It is possible to roughly estimate how many amino acids are present in these disordered regions, allowing for a semi-quantitative, accurate description of the dynamic structure of IDPs.
References and Further Reading
Sivarajah, I. (2021) Atomic Force Microscopy: General Principles and Applications. [Online] AZoOptics.com. Available at: https://www.azooptics.com/Article.aspx?ArticleID=2083
Kodera, N., Noshiro, D., Dora, S.K. et al. (2021) Structural and dynamics analysis of intrinsically disordered proteins by high-speed atomic force microscopy. Nat. Nanotechnol. 16, 181–189 https://doi.org/10.1038/s41565-020-00798-9
Shingo Fukuda and Toshio Ando. (2021) Faster high-speed atomic force microscopy for imaging of biomolecular processes. Review of Scientific Instruments 92, 033705 (2021) https://doi.org/10.1063/5.0032948
Disclaimer: The views expressed here are those of the author expressed in their private capacity and do not necessarily represent the views of AZoM.com Limited T/A AZoNetwork the owner and operator of this website. This disclaimer forms part of the Terms and conditions of use of this website.