Apr 16 2015
A device developed by Massachusetts General Hospital investigators may bring rapid, accurate molecular diagnosis of tumors and other diseases to locations lacking the latest medical technology. In their report appearing in PNAS Early Edition, the researchers describe a smartphone-based device that uses the kind of technology used to make holograms to collect detailed microscopic images for digital analysis of the molecular composition of cells and tissues.
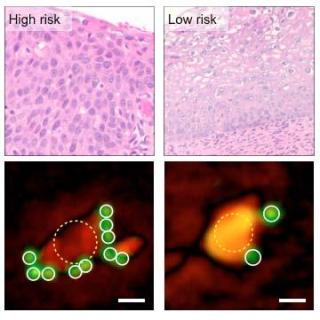 By quantifying the number of tumor-marker-targeting microbeads bound to cells (lower images), the D3 system categorizes high- and low-risk cervical biopsy samples as well as traditional pathology (upper images). Credit: Massachusetts General Hospital Center for Systems Biology
By quantifying the number of tumor-marker-targeting microbeads bound to cells (lower images), the D3 system categorizes high- and low-risk cervical biopsy samples as well as traditional pathology (upper images). Credit: Massachusetts General Hospital Center for Systems Biology
"The global burden of cancer, limited access to prompt pathology services in many regions and emerging cell profiling technologies increase the need for low-cost, portable and rapid diagnostic approaches that can be delivered at the point of care," says Cesar Castro, MD, of the MGH Cancer Center and Center for Systems Biology, co-lead author of the report. "The emerging genomic and biological data for various cancers, which can be essential to choosing the most appropriate therapy, supports the need for molecular profiling strategies that are more accessible to providers, clinical investigators and patients; and we believe the platform we have developed provides essential features at an extraordinary low cost."
The device the team has developed - called the D3 (digital diffraction diagnosis) system - features an imaging module with a battery-powered LED light clipped onto a standard smartphone that records high-resolution imaging data with its camera. With a much greater field of view than traditional microscopy, the D3 system is capable of recording data on more than 100,000 cells from a blood or tissue sample in a single image. The data can then be transmitted for analysis to a remote graphic-processing server via a secure, encrypted cloud service, and the results rapidly returned to the point of care.
For molecular analysis of tumors, a sample of blood or tissue is labeled with microbeads that bind to known cancer-related molecules and loaded into the D3 imaging module. After the image is recorded and data transmitted to the server, the presence of specific molecules is detected by analyzing the diffraction patterns generated by the microbeads. The use of variously sized or coated beads may offer unique diffraction signatures to facilitate detection. A numerical algorithm developed by the research team for the D3 platform is capable of distinguishing cells from beads and of analyzing as much as 10 MB of data in less than nine hundredths of a second.
A pilot test of the system with cancer cell lines detected the presence of tumor proteins with an accuracy matching that of the current gold standard for molecular profiling, and the larger field of view enabled simultaneous analysis of more than 100,000 cells at a time. The investigators then conducted analysis of cervical biopsy samples from 25 women with abnormal PAP smears - samples collected along with those used for clinical diagnosis - using microbeads tagged with antibodies against three published markers of cervical cancer. Based on the number of antibody-tagged microbeads binding to cells, D3 analysis promptly and reliably categorized biopsy samples as high-risk, low-risk or benign, with results matching those of conventional pathologic analysis.
D3 analysis of fine-needle lymph node biopsy samples was accurately able to differentiate four patients whose lymphoma diagnosis was confirmed by conventional pathology from another four with benign lymph node enlargement. Along with protein analyses, the system was enhanced to successfully detect DNA - in this instance from human papilloma virus - with great sensitivity. In these pilot tests, results of the D3 assay were available in under an hour and at a cost of $1.80 per assay, a price that would be expected to drop with further refinement of the system.
"We expect that the D3 platform will enhance the breadth and depth of cancer screening in a way that is feasible and sustainable for resource limited-settings," says Ralph Weissleder, MD, PhD, director of the MGH Center for Systems Biology (CSB) and co-senior author of the paper. "By taking advantage of the increased penetration of mobile phone technology worldwide, the system should allow the prompt triaging of suspicious or high-risk cases that could help to offset delays caused by limited pathology services in those regions and reduce the need for patients to return for follow-up care, which is often challenging for them."
In their further development of this technology, co-senior author Hakho Lee, PhD, notes, "The research team will: 1) investigate the D3 platform's ability to analyze protein and DNA markers of other disease catalysts, including infectious agents and allergens; 2) integrate the software with larger databases; and 3) conduct clinical studies in settings such as care-delivery sites in developing countries or rural settings and for home testing with seamless sharing of information with providers and/or clinical investigators." The MGH has filed a patent application covering the D3 technology.
Source: http://www.mgh.harvard.edu